Paula Oset, Simon Claus, Klaas Deneudt, Elisabeth Debusschere
An important objective of JERICO-Next has been to promote a stronger integration of biological data within the observation networks in order to address pelagic and benthic biodiversity questions. This biological information can be gathered with established methods but also with innovative observation techniques capable of delivering operational (near real-time) data. Part of the work package on data management (WP5) in JERICO-Next focuses on making sure that the biological (meta)data collected through the project aligns with the international standards that make possible for the data to flow to the European Data Infrastructures.
During the recent years, large marine biological data systems have been created to store, archive and integrate traditional marine biological data, e.g. (Eur)OBIS, EMODnet Biology. In WP5 we described the general data management practices, data standards (Darwin Core) and quality check procedures that are currently applied for biodiversity data in these European Data Infrastructures. We have also inventoried the different data types that will result from the JERICO-Next project, making a distinction between the more mature ones and those data collected with emerging technologies. The current standards can be easily applied to the pelagic or benthic data collected with traditional sampling methods. However, some data derived from developing technologies and sensors was not fully ready to be ingested by the existing marine biological data networks in an operational way.
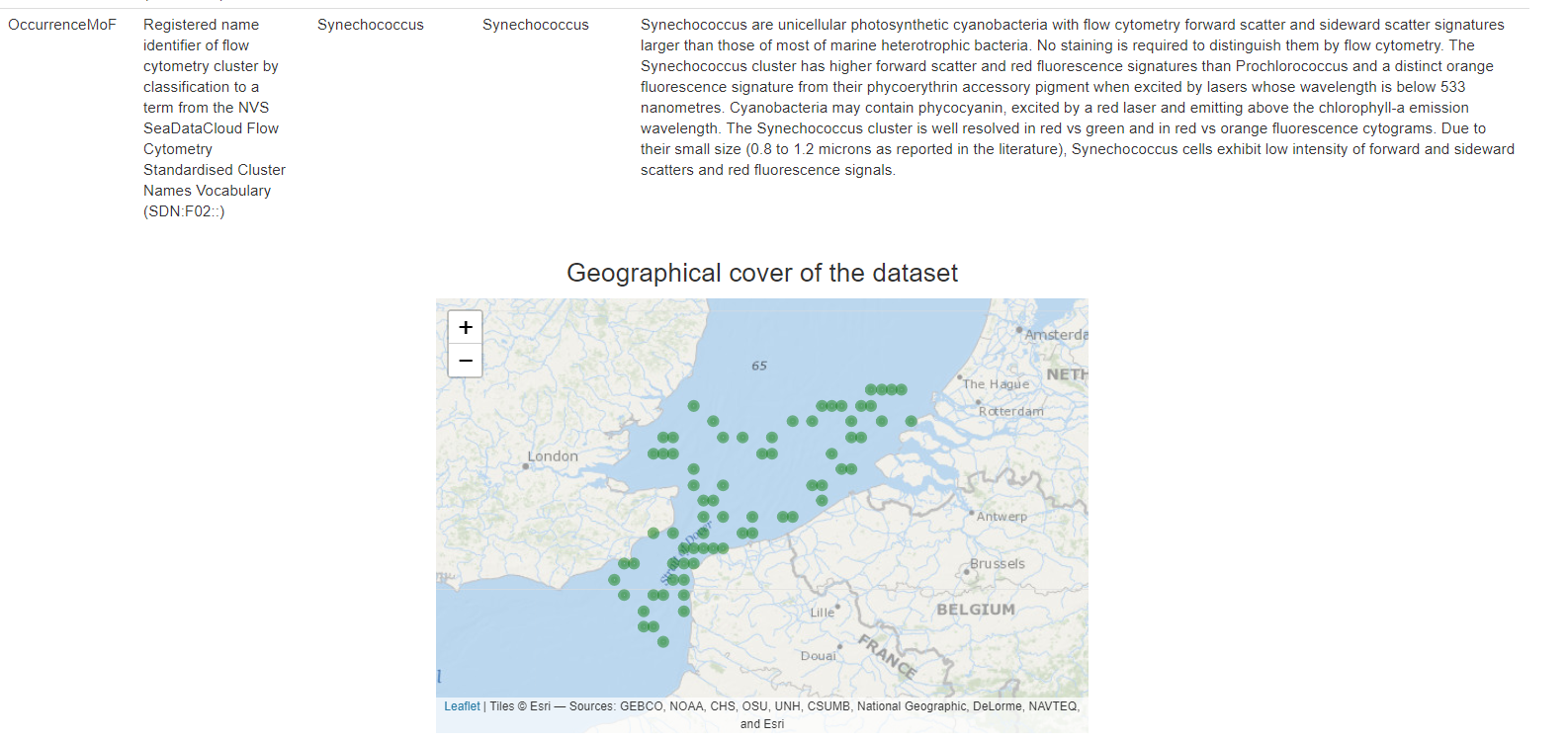
Taking advantage of synergies with complementary projects, we have explored the possibility to adapt and expand the current data schemas in order to facilitate the integration of this novel data on pelagic and benthic biodiversity. For example, the SeaDataCloud project holds a specific task to work on the ingestion, validation, long-term storage and access of Flow Cytometer (FCM) data. In this context, new controlled vocabularies have been developed by the FCM community to store the cluster and optical properties data from FCM observations. Besides, a transition from Darwin Core Occurrence to the Darwin Core Event schema has recently been implemented by (Eur)OBIS and EMODnet Biology, allowing for more flexibility and the possibility to accommodate additional data types.
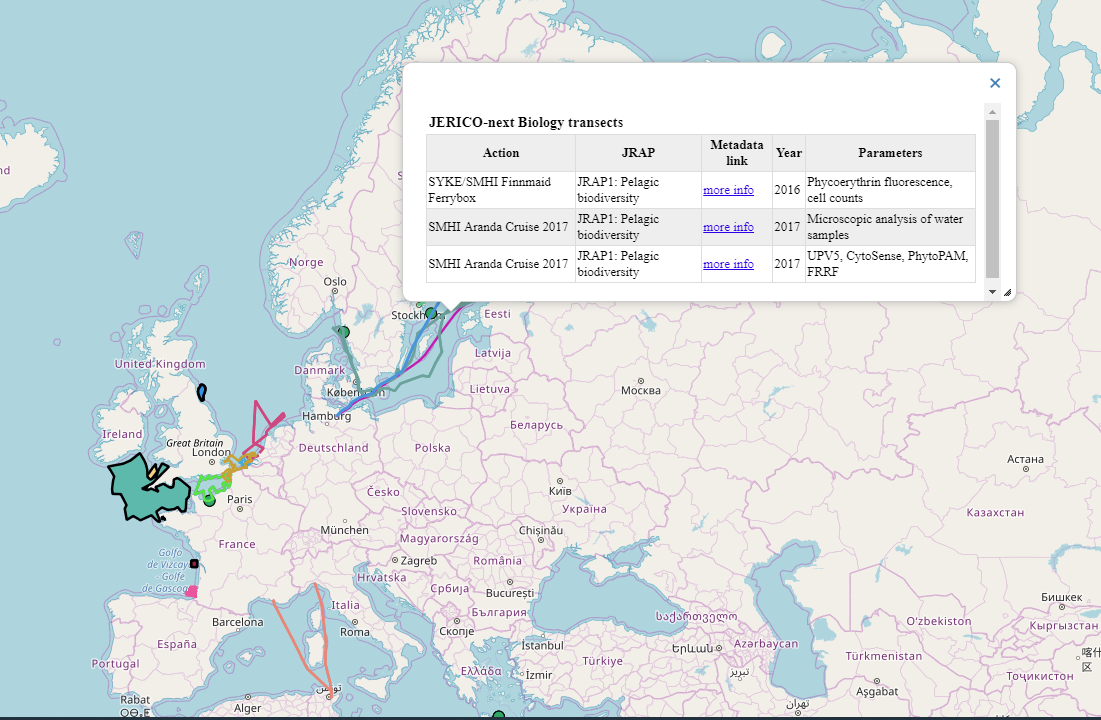
A description of biological data collected throughout the project can be found at the Data access section of JERICO-Next website. A metadata record is available for each dataset in the EMODnet Biology catalogue, where the characteristics, state and accessibility and terms of use of the data are documented. These metadata records can also be accessed using a map interface where the geographical scope of each dataset is displayed. Once a dataset is fully processed, harmonized and QCed, direct access to the data is provided in the metadata record, where a link to an archived version of the raw data might also be. Once the data is integrated in EMODnet Biology, it is also findable using the data download toolbox. This way, we facilitate the exchange of the data generated by the project between different users, including the project’s partners.